Brain tumors are a particularly deadly form of cancer, and the leading cause of cancer-related death in children. Two studies published this week in Nature journals applied next-generation sequencing to pediatric brain tumors, revealing a striking pattern of recurrent somatic mutations in H3F3A, a gene encoding the histone prorein H3.3. These are the first unbiased surveys of childhood brain cancer, and also the first reports of driver mutations in histone H3.
Brain Cancer in Adults and Children
About 22,000 new cases of brain and central nervous system (CNS) tumors are diagnosed each year in the United States, and 3,000 of those are in children. Both incidence and mortality are higher in Caucasians than other racial/ethnic groups.
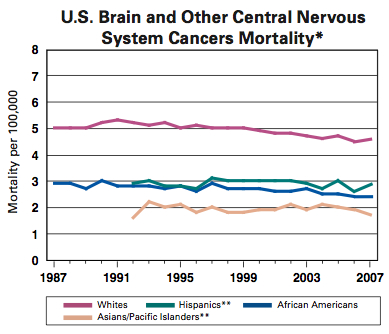
Source: NCI
Across all ethnicities, men have higher incidence and mortality than women. And though brain cancers are not listed among the ?common? cancer types, about $3.5 billion is spent annually treating them in the United States.
Glioma and Glioblastoma Multiforme
Brain tumors are the most prevalent of any pediatric cancer (27%), and certainly among the more lethal. One such cancer, pediatric diffuse intrinsic pontine glioma (DIPG), arises almost exclusively in children and has a long-term survival of less than 10%. Glioblastoma multiforme (GBM), an aggressive brain tumor affecting both adults and children, has dismal outcomes as well:
Most patients die within a few years of diagnosis, despite aggressive therapy. Ted Kennedy died of it. GBM is less common in the pediatric setting, but the primary tumors that are diagnosed in children arise de novo and are morphologically indistinguishable for adult GBM. While the Cancer Genome Atlas consortium characterized several thousand genes in adult GBM, this disease is understudied in the pediatric setting.
Exome Sequencing in Pediatric Glioblastoma
To better understand this disease, Schwartzenruber et al performed exome sequencing (they call it ?whole-exome? sequencing, but we know better) on 48 well-characterized pediatric GBMs, 6 of which had matched normal DNA allowing identification of somatic mutations. Using Illumina?s TruSeq exome kit and HiSeq 2000 platform, the authors generated ~13.4 Gbp of data per sample, achieving 91% of target bases covered at least 10x, with an average depth of 61x.
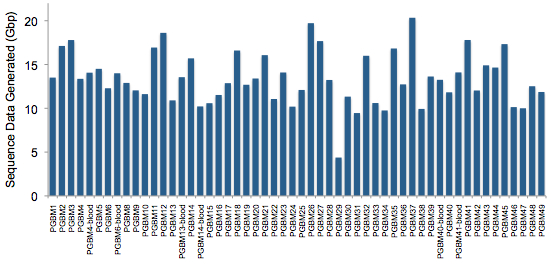
The six tumors with matched normal DNA harbored 15 somatic mutations on average (range 3-31), a mutation rate lower than adult GBM but higher than another pediatric brain tumor, medulloblastoma.
Recurrent Mutations in H3F3A
Four of the six cases had heterozygous mutations in the H3F3A gene, two at K27M and two at G34R. This gene encodes the replication-independent histone variant H3.3. To find two recurrent mutations in four unrelated cases is quite striking, particularly because both occurred in the amino-tail of the protein at positions believed to undergo repressing or activating post-translational modifications. Extending the analysis to the 42 other exomes, the authors found that:
- 31% of tumors (15/48) had mutations at residue 27 or 34 in H3F3A.
- 31% of tumors (15/48, including 8 of the above) had mutations in chromatin remodeling genes ATRX/DAXX.
- 54% of cases had somatic TP53 mutations. Among samples with H3F3A/ATRX/DAXX mutations, this frequency jumped to 86%.
Whole-genome Sequencing of Pediatric Glioma
 Also this week, members of the St. Jude Children?s Research Hospital ? Washington University Pediatric Cancer Genome Project (PCGP) described the whole-genome sequencing pediatric diffuse intrinsic pontine glioma (DIPG), 7 cases with tumor and matched germline DNA. Writing in Nature Genetics, Wu et al reported that 4 0f the 7 cases harbored the K27M mutation in H3F3A. They next undertook targeted sequencing of the lysine-27 residue in all 16 genes that code for H3 isoforms in a validation cohort comprising 43? DIPGs and 36 non-brainstem glioblastomas.
Also this week, members of the St. Jude Children?s Research Hospital ? Washington University Pediatric Cancer Genome Project (PCGP) described the whole-genome sequencing pediatric diffuse intrinsic pontine glioma (DIPG), 7 cases with tumor and matched germline DNA. Writing in Nature Genetics, Wu et al reported that 4 0f the 7 cases harbored the K27M mutation in H3F3A. They next undertook targeted sequencing of the lysine-27 residue in all 16 genes that code for H3 isoforms in a validation cohort comprising 43? DIPGs and 36 non-brainstem glioblastomas.
They found striking recurrence of lysine-27 mutations in H3F3A or the closely related gene HIST1H3B (which encodes H3.1) in 78% of gliomas and 22% of non-brainstem glioblastomas. They also identified the second recurrent mutation (Gly34Arg) in 14% of the glioblastomas, but none of the gliomas, findings remarkably consistent with the GBM study. Screening of several other pediatric brain tumors revealed that the histone H3 mutations seem exclusive to pediatric high-grade gliomas.
Histone H3 and Glioma Pathogenesis
To my knowledge, this is the first report of somatic mutations in histone H3. The isoforms of this histone play complex regulatory roles in several cellular processes ? epigenetic regulation of gene expression, selective regulation of developmental genes, and telomere maintenance ? that could contributed to tumor growth and progression if dysregulated. Indeed, the precise location of these recurrent events at two specific residues, both of which are at or near regulatory post-translation modification positions, suggest a gain-of-function. If true, then the mutated forms of these genes may offer new targets for the surveillance and/or treatment of this lethal disease.
References
Schwartzentruber J, Korshunov A, Liu XY, et al (2012). Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature PMID: 22286061
St. Jude Children?s Research Hospital?Washington University Pediatric Cancer Genome Project (2012). Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature genetics PMID: 22286216
Source: http://www.massgenomics.org/2012/02/recurrent-histone-alterations-in-pediatric-brain-cancer.html
drop dead gorgeous 2011 nfl playoff schedule cowboys vs giants ndaa dallas cowboys weight watchers timberwolves
No comments:
Post a Comment
Note: Only a member of this blog may post a comment.